Structural Network Analysis
Biochemical reactions are characterized by their participants, the participants’ stoichiometries, reaction directions, and catalyzing enzymes. They are the constituent elements of any cellular function. The structures of biochemical reaction networks determine to some extent, which functions they can establish. In addition, these structures are often well-characterized, for example, for metabolic networks. We aim to increase and exploit knowledge on network structures to predict network functions, which requires novel models and analysis methods.
Metabolic Network Reconstruction
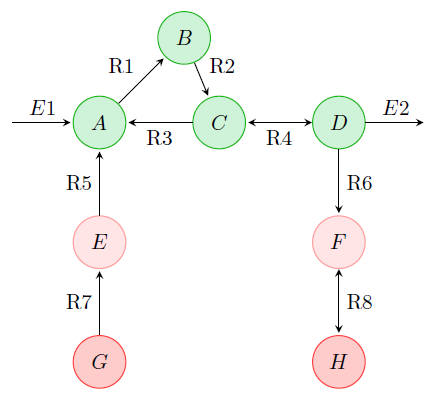
Genome-scale metabolic networks are a specific, widely used class of structural models. However, model development is so far largely manual, and therefore tedious and error-prone. In the external page SystemsX.ch project MetaNetX, we aim to automate this process and to generate experimentally testable hypotheses on new metabolic reactions. For example, we developed a conceptually novel computational method based on probabilistic models over network patterns to infer features of metabolic reactions and networks such as reaction directions and enzyme classes. In collaboration with the SIB-Swiss Institute of Bioinformatics, we make the resulting, standardized models as well as the methods for metabolic network construction and analysis available via the web site external page www.metanetx.org.
Ganter M, Kaltenbach HM, Stelling J (2014) Predicting network functions with nested patterns. Nat Commun 5: 3006. external page http://doi.org/10.1038/ncomms4006.
Ganter M, Bernard T, Moretti S, Stelling J, Pagni M (2013) MetaNetX.org: a website and repository for accessing, analysing and manipulating metabolic networks. Bioinformatics 29: 815-816. external page http://doi.org/10.1093/bioinformatics/btt036.
Terzer M, Maynard ND, Covert MW, Stelling J (2009) Genome-scale metabolic networks. Wiley Interdisciplinary Reviews-Systems Biology and Medicine 1: 285-297. external page http://doi.org/10.1002/Wsbm.37.
Metabolic Pathways
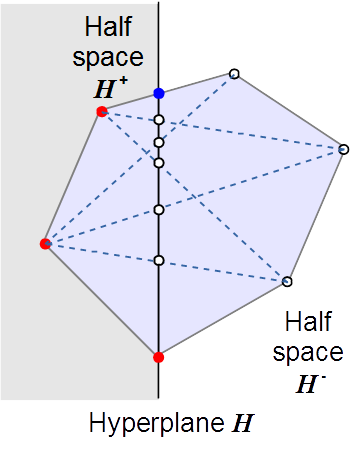
Formally defined metabolic pathways – so-called elementary flux modes (EFMs) – provide means of characterizing all feasible fluxes in a network. They have close relations to concepts in computational geometry and mathematical optimization, but determining all EFMs for a given network is computationally hard. We have developed efficient algorithms for the computation of EFMs that are freely available as software tools efmtool and polco. Applications to yeast metabolism, for example, revealed how the interplay of network structure and thermodynamics limits the organism’s metabolic network operation across compartment boundaries.
Jol SJ, Kummel A, Terzer M, Stelling J, Heinemann M (2012) System-level insights into yeast metabolism by thermodynamic analysis of elementary flux modes. PLoS Comput Biol 8: e1002415. external page http://doi.org/10.1371/journal.pcbi.1002415.
Terzer M, Stelling J (2008) Large-scale computation of elementary flux modes with bit pattern trees. Bioinformatics 24: 2229-2235. external page http://doi.org/10.1093/bioinformatics/btn401.
Klamt S, Stelling J (2003) Two approaches for metabolic pathway analysis? Trends Biotechnol 21: 64-69. external page http://doi.org/10.1016/S0167-7799(02)00034-3.
Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED (2002) Metabolic network structure determines key aspects of functionality and regulation. Nature 420: 190-193. external page http://doi.org/10.1038/nature01166.
Metabolism and Regulation
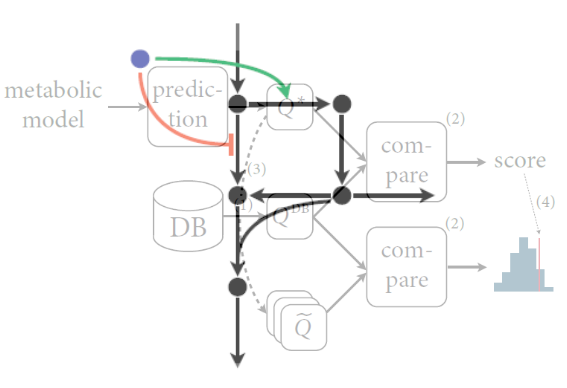
When network structures impose constraints on metabolic network operation, then they should also pose constraints on how metabolism reacts to perturbations and on how metabolic regulation should be designed. Based on this idea, we developed methods to infer the likely (optimal) control structure for metabolism, and to use these concepts to integrate experimental data from different levels of cellular organization. For example, we used our approach of structural sensitivity analysis to determine when the bacterium Bacillus subtilis uses metabolic or transcriptional regulation to adapt to dynamically changing nutrients.
Chubukov V, Uhr M, Le Chat L, Kleijn RJ, Jules M, et al. (2013) Transcriptional regulation is insufficient to explain substrate-induced flux changes in Bacillus subtilis. Mol Syst Biol 9: 709. external page http://doi.org/10.1038/msb.2013.66.
Buescher JM, Liebermeister W, Jules M, Uhr M, Muntel J, et al. (2012) Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335: 1099-1103. external page http://doi.org/10.1126/science.1206871.
Uhr M, Villaverde AF, Egea JA, Banga JR, Stelling J (2011) Inference of transcriptional control design of metabolic networks. Proceedings of the 18th IFAC World Congress. pp. 10448-10453. external page http://doi.org/10.3182/20110828-6-IT-1002.01588.
Uhr M, Stelling J (2008) Structural sensitivity analysis of metabolic networks. Proc 17th World Congress, The International Federation of Automatic Control; Seoul, Korea, July 6-11, 2008. IFAC. pp. 15879 - 15884. external page http://doi.org/10.3182/20080706-5-KR-1001.02684.
Structure, Modules, and Dynamics
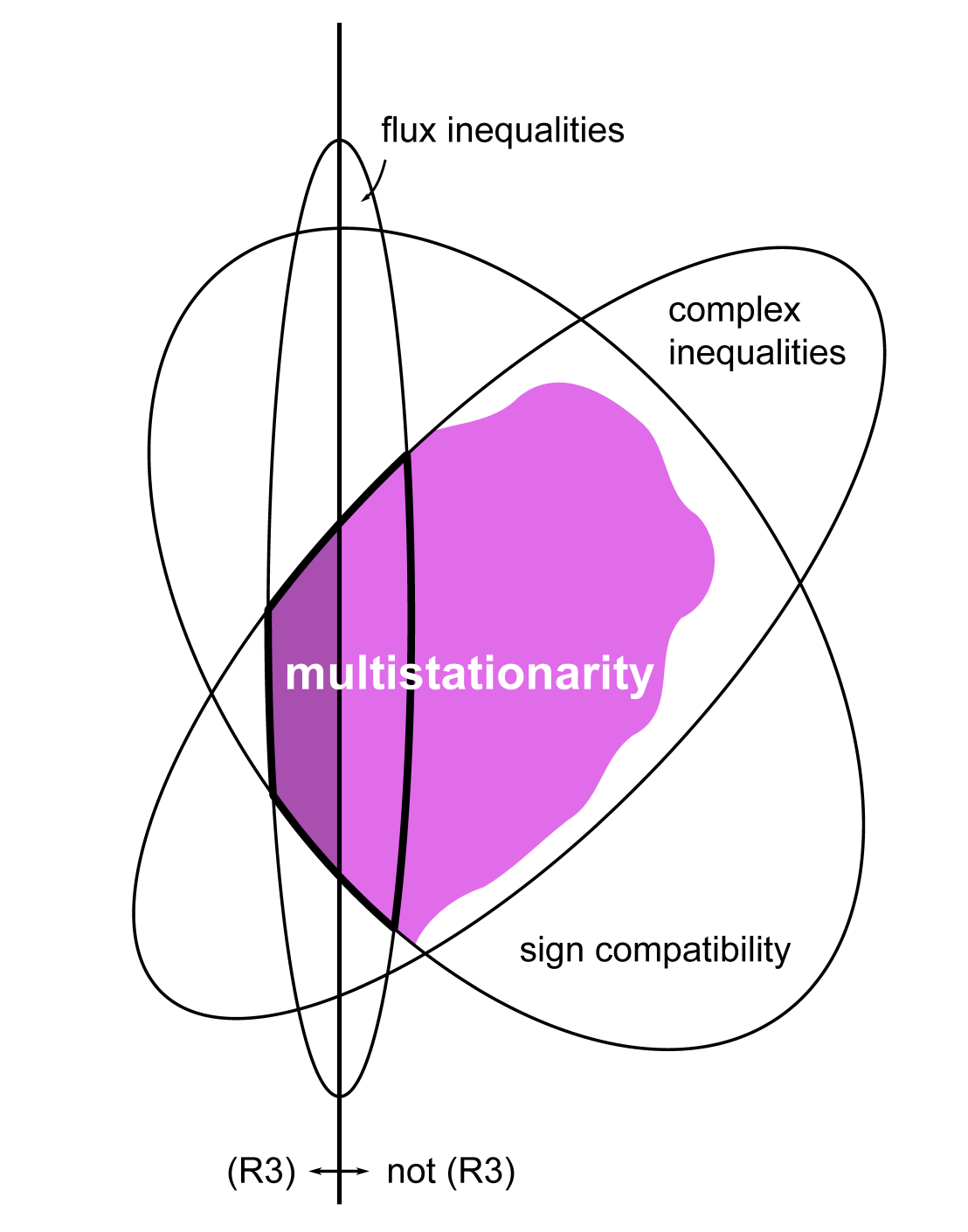
Network structures also constrain the possible dynamics of a biological system, and one does not need to know parameter values to determine certain aspects of a network’s qualitative behavior. Structural analysis methods of this type therefore have close links to, and help in dynamic systems analysis. We are developing analysis methods to decompose complex networks into smaller modular, such as to make, for example, parameter estimation in large networks feasible. We also extend Chemical Reaction Network Theory (CRNT), a parameter-free approach to determine dynamic network properties such as multistationarity. We proposed, for example, the application of CRNT to EFMs for a modular network analysis.
Lang M, Summers S, Stelling J (2014) Cutting the wires: modularization of cellular networks for experimental design. Biophys J 106: 321-331. external page http://doi.org/10.1016/j.bpj.2013.11.2960.
Otero Muras I, Yordanov P, Stelling J (2012) Advances in chemical reaction network theory for the identification of kinetic models. System Identification. pp. 1713-1718. external page http://doi.org/10.3182/20120711-3-BE-2027.00399.
Kaltenbach HM, Stelling J (2012) Modular analysis of biological networks. Adv Exp Med Biol 736: 3-17. external page http://doi.org/10.1007/978-1-4419-7210-1_1.
Kaltenbach H-M, Constantinescu S, Feigelman J, Stelling J (2011) Graph-based decomposition of biochemical reaction networks into monotone subsystems. Algorithms in Bioinformatics: Springer Berlin Heidelberg. pp. 139-150. external page http://doi.org/10.1007/978-3-642-23038-7_13.
Conradi C, Flockerzi D, Raisch J, Stelling J (2007) Subnetwork analysis reveals dynamic features of complex (bio)chemical networks. Proc Natl Acad Sci U S A 104: 19175-19180. external page http://doi.org/10.1073/pnas.0705731104.