Experimental Yeast Biology
Fission (Schizosaccharomyces pombe) and budding (Saccharomyces cerevisiae) yeast, our model systems, are simple, single cell eukaryotes. They are among the best understood model systems, easy to manipulate, and a wide variety of tools is available or are under development. We aim to understand how a network performs its function in a cellular setting. We focus on metabolism and its interactions with, and effects on the (physiological) state of the cell. We study these phenomena from diverse angles, including synthetic biology approaches, to ultimately understand, for example, whether a network’s short term behavior is predictive for its long term function.
Quantitative Signaling
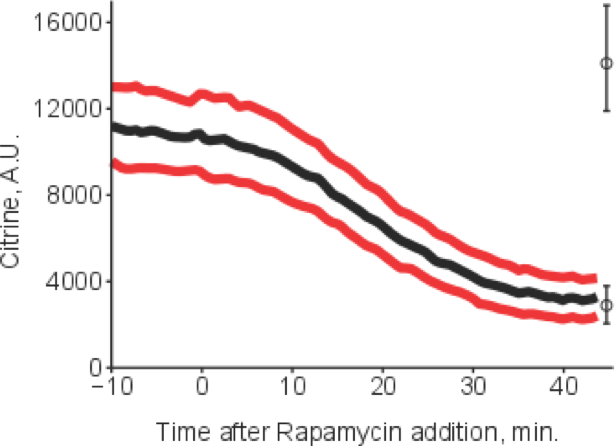
Any cell needs to monitor its environment and adapt to changes therein. In more abstract terms, signals arise, they are sensed, transduced and interpreted. We aim to quantitatively understand the signals arising from metabolism, their sensing and transduction, and how they ultimately lead to changes in the physiological state of the cell. We try to tackle these problems by combining quantitative single cell measurements and dynamic mathematical modelling. In particular, we precisely measure crucial points of regulation in a variety of changing conditions and/or genetic perturbations. This allows tight interactions between experiments and models, for example for cross-talk in yeast stress signaling via Msn2 as a problem we currently address.
To come soon.
Synthetic Biology
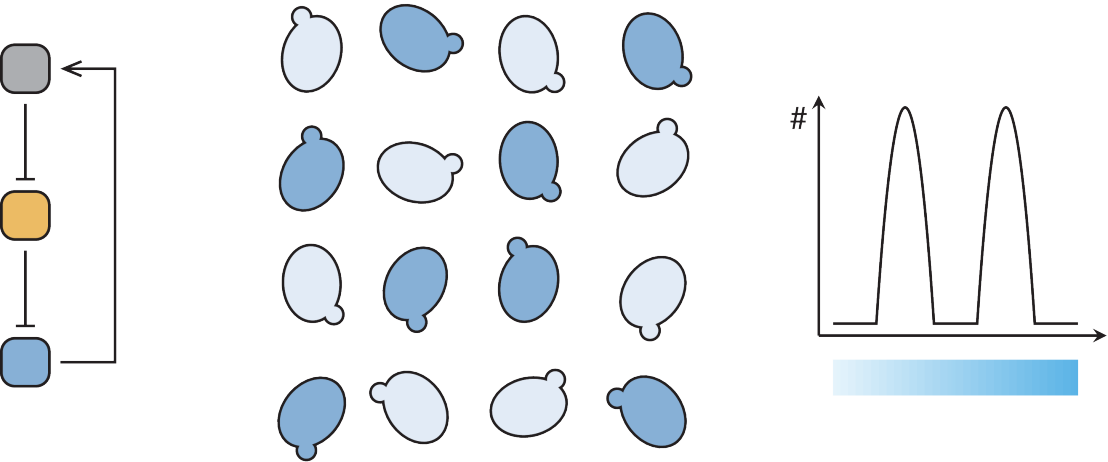
We aim to rationally design and create yeast systems able to perform a function of choice. For example, an engineered yeast cell expressing a set of mammalian receptors could sense and integrate signals of different origin. It could be used to study signal transduction and crosstalk, to engineer metabolic regulation into signaling pathways, or just to create efficient readouts to screen small molecules specifically modulating a single signal transduction pathway. Rational design of complex functions is an ambitious goal. We started by characterizing proteins performing basic biological function such as inducing or repressing transcription, controlling their subcellular localization and their half-lives. We assembled small circuits with desired properties such as combining two signals or creating a bistable gene network and analyzed their quantitative behavior and interaction with the host cell.
Marchisio MA, Rudolf F (2011) Synthetic biosensing systems. Int J Biochem Cell Biol 43: 310-319. external page http://doi.org/10.1016/j.biocel.2010.11.012
Enabling Technologies: Wetware
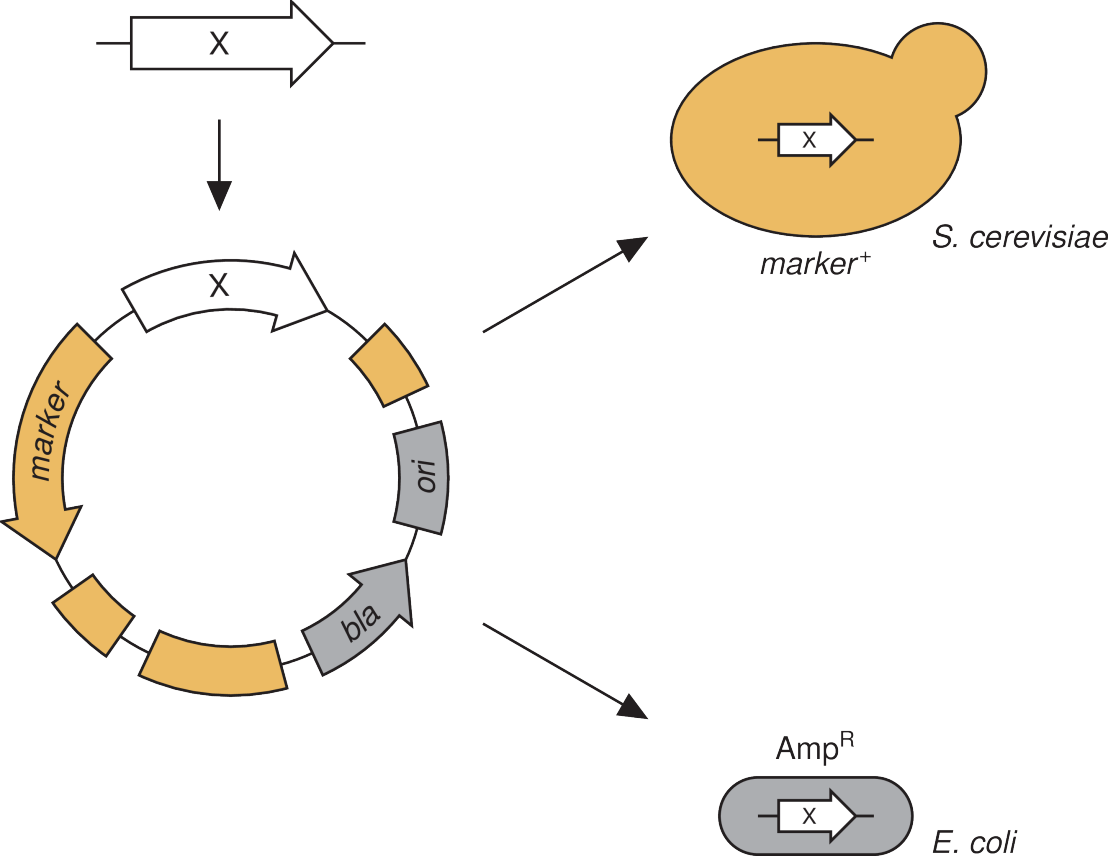
The quantitative study of biological system requires means to precisely manipulate cells and sensors to faithfully report events. A plethora of tools has been developed allowing the precise interrogation of single yeast cells. We aim to complement these methods by refining them into broadly applicable tools or devising ways to more precisely monitor cellular concentrations and conditions. For example, we developed an improved version of a hormone inducible transcription factor. While many transcription factors are available, most depend on the growth conditions and/or are toxic upon induction. By combining experiment and theory, we implemented a broadly applicable, non-toxic transcription factor which, in combination with a series of engineered promoters, allows a tight control of gene expression. Similar efforts for precise manipulation of the host genome and for regulation of protein levels are ongoing.
Ottoz DS, Rudolf F, Stelling J (2014) Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae. Nucleic Acids Res 42: e130. external page http://doi.org/10.1093/nar/gku616
Enabling Technologies: Hardware
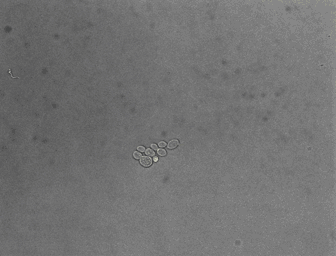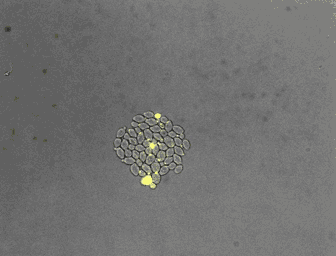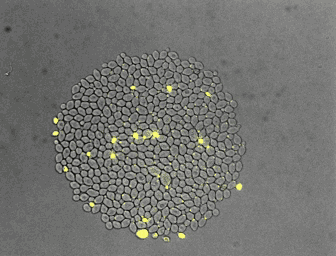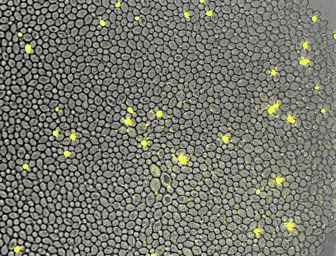
For the quantitative analysis of cells and their behavior, we often use fluorescence microscopy. Microfluidics enables a precise control of the environment and can facilitate the acquisition of high resolution images. In collaboration with the Hierlemann group, we developed a series of platforms allowing cells to grow for a long time in constant or changing environments. Manipulation and recording of cells are tedious, labor intensive processes. With laboratory automation, we strive to facilitate experimental setup, strain construction, or pipetting. Further, we constantly incorporate technologies in our analysis platform to increase the number of parameters obtainable from single cells.
Zhu Z, Frey O, Ottoz DS, Rudolf F, Hierlemann A (2012) Microfluidic single-cell cultivation chip with controllable immobilization and selective release of yeast cells. Lab Chip 12: 906-915. external page http://doi.org/10.1039/c2lc20911j